The Current State of Nutrigenomics and Personalized Nutrition
As a scientist my job is to be objective, and pull no punches. It also requires a great deal of self-punches. To quote the great Richard Feynman, “The first principle is that you must not fool yourself and you are the easiest person to fool”.
Let me begin by laying the landscape. Personalized medicine, which includes things like nutrigenomics, is undoubtedly the future. The evidence is overwhelming that each person has a unique “molecular fingerprint” and that the future of health will be defined by personalized, dense, dynamic data clouds that includes virtually every quantifiable data point about you.
That is the future, that is not the present. We are merely scratching the surface and our understanding is currently very young and naive, especially when we only considered genomic data (i.e. data that comes from only your DNA). To fully explain what we are looking at and understand how genetic information can be interpreted, specifically with regard to nutrigenomics, we need to move to another landscape, how biology truly works.
It Isn’t All About the Genome
To fully understand how one’s genome can impact things like nutrition, performance, and recovery, one needs to be aware of the fact that the genome does not operate in isolation. For decades we have be given a crude analogy that the genome is a blueprint and conveys all the information needed and that one gene equals one protein.
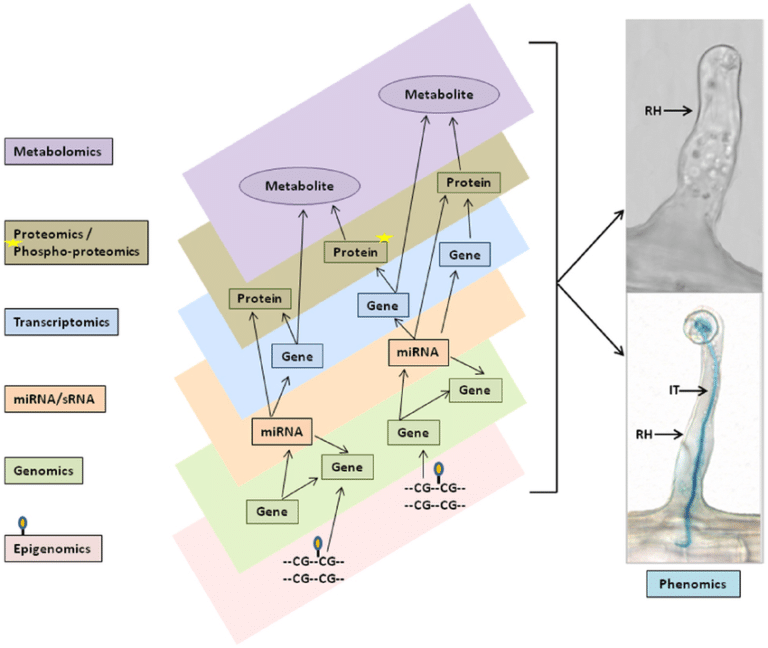
The past two decades have shown us that there are levels of “regulation” that mediate how a genome impacts ones phenotype (e.g. metabolism, performance, recovery, etc.).
We are just beginning to scratch the surface of the regulatory mechanisms and we are finding that the genome is one piece, albeit an important piece, in a multilayer system that turns underlying digital information (i.e. the genome) into the complex organism that is a human.
Before we even dive into the current state of nutrigenomics and performigenomics (I believe this is the first use of this term in the literature) a brief depiction of these layers of controls nicely highlights why simple alterations in the genome that might associate with a phenotype (more on this later) is currently a vast oversimplification.
The use of genomic information to identity a phenotype falls on a spectrum. On one end of the spectrum you have things like monogenic disease such as cystic fibrosis, tay-sachs, and phenylketonuria. These diseases are the result of a mutation in a single gene and the resulting phenotype is essentially binary. The underlying cause of these diseases is a substantial “problem” with a protein that serves an important biological function.
On the other end of the spectrum you have phenotypes that are controlled by interactions of many genes, sometimes 10 genes, sometimes 1000 genes. Things that are regulated by many genes such as muscle metabolism, muscle fiber type and function, and response to mechanical (e.g. resistance training) and chemical (e.g. lactate accumulation) are very unlikely to be able to be linked single mutations.
What You Need to Know About The Current State of Nutrigenomics
There are a few key technical things to know about current nutrigenomics and performigenomics that can substantially improve your understanding and what the results of tests like these mean. They are mostly statistical in nature but also biological. Let us cover the statistical first.
Current nutrigenomic and performigenomic platforms are based on a technology that measure simple point mutations known as single nucleotide polymorphisms (SNPs). Essentially this is as simple as a single DNA base pair changing from an A to a T, C, or G. These single changes in each gene are then associated with a certain phenotype.
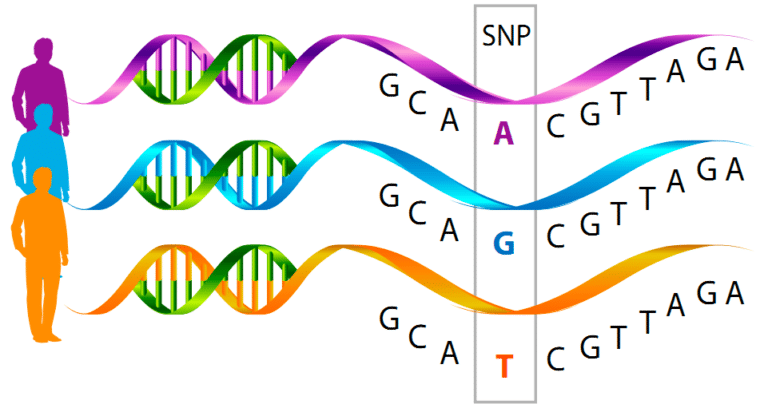
In most of the current tests these SNPs are associated with a phenotype and the explain a very small portion of the variance, often times 5-10%. What this means is that if you take 5000 people with an A and 5000 people with a B, that “genetic mark” can only explain the phenotype of about 50-100 of those people. That means there are other things that explain 90-95% of the phenotype.
Let me put this in context. If you get a genetic result saying that your SNP says you should consume more carbohydrates than fats and it explains 10% of the variance seem in humans, that means 90% of the reason you select your diet is likely not due to this variant; things like your environment, training, and goals play a much bigger role in deterring how your body optimally processes.
The other key statistical point to understand about many of these tests relates to risk of disease and understanding the idea of chance versus determinism. There are genetic mutations that lead directly to diseases (e.g. Downs’ Syndrome, Hemophilia, Tay-Sach’s, etc.). As mentioned earlier these mutations always cause disease.
There are other genetic markers that do not mean you get that disease, but they increase your risk (e.g. your probability of getting a disease). In practice, this means that if you get a result that tells you that you have an increased risk of Alzheimer’s it is not a diagnosis, it is simply an increased risk. For example, if the baseline risk of Alzheimer’s is 20% by the age of 70 and you have a result that tells you there is a 40% increased risk of getting Alzheimers that means you have a 28% total risk (20% X 1.40=28%), not at 40% risk.
The Current Value of Nutrigenomics and Performigenomic Tests
Currently, almost all results that apply to nutrition that come from SNP analysis fall under the two following categories: 1) Complex Phenotypes, 2) Low Explanatory value. When we look at carbohydrate, fat, and protein metabolism and their role on both overall health and athletic performance, it is abundantly clear that these are complex phenotypes and SNP data can not unpack these complexities. Additionally, these SNP analysis are often of low explanatory value, meaning objectively they explain a very small amount of the difference in phenotypes (usually <10%).
Conclusion
The current state of nutrigenomics and performigenomics is nascent and while personalized nutrition and training programming that can be based on biological data is the future we are not quite there. The genome is but one piece of what regulates our biology, we need much richer, more complex data. Use these analysis as hypothesis generating tools, not rigid guides on how to program your own nutrition and training.